Homodimer and heterodimer interfaces
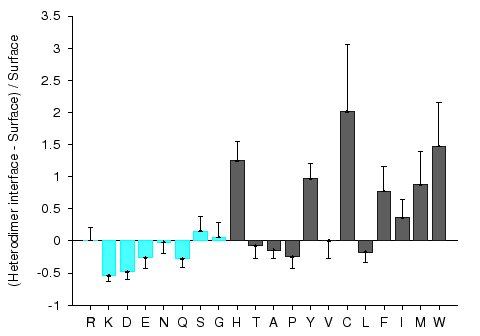
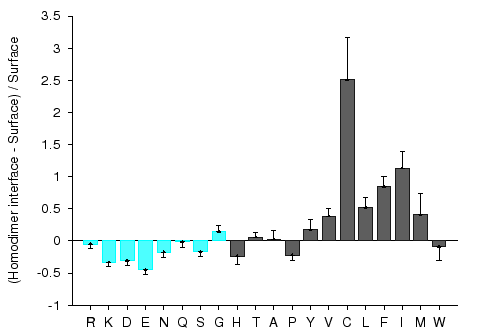
Protein-protein interaction sites have been intensively studied in an attempt to understand the molecular determinants of protein recognition and to identify specific characteristics of the interactions, such as residue propensities, residue pairing preferences, hydrophobicity, size, shape, solvent accessibility, and hydrogen bond protection. Homocomplexes, for example, are often permanent and optimized, whereas many heterocomplexes are nonobligatory, associating and disassociating according to the environmental or external factors and involve proteins that must also exist independently (Jones and Thornton, 1996).
Figures 1 and 2 give composition profiles of interface residues of homodimers and heterodimers in comparison to the amino acid composition of surfaces of monomeric proteins. Both kinds of interfaces are generally enriched in hydrophobic residues (right hand side of the graph), which in part explains their propensity towards complexation. Interfaces of heterodimers are enriched in polar histidine and tyrosine, which is consistent with the finding that transient protein-protein complex interfaces are more polar than those of stable oligomeric proteins (Janin et al. 1988; Jones and Thornton, 1996; Valdar and Thornton 2001). Heterocomplex interfaces are enriched in all three major aromatics (trypthopan, tyrosine, and phenylalanine), as these three residues are bulky, planar and rigid which enhances the prospects for binding.
α-MoRF Composition
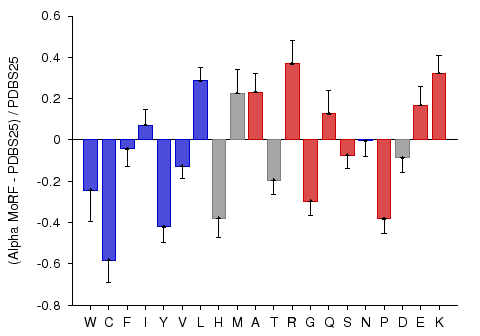
According to the MoRF hypothesis, MoRFs are disordered in the absence of binding partners (Oldfield et al., 2005; Mohan et al., 2006), and consequently their amino acid compositional biases may be more similar to intrinsically disordered proteins than to ordered proteins. Alternatively, the binding propensities of these regions may give the compositions of these proteins a bias somewhere in between those of ordered and disordered proteins. To test the compositional bias of MoRFs, the fractional difference between MoRF compositions and PDBS25 compositions was calculated. This analysis revelaed that that MoRFs are enriched in many of the disorder-promoting amino acids, arginine, glycine, serine, and proline, and depleted in many of the order promoting amino acids, tryptophan, isoleucine, tyrosine, valine, and leucine. These biases suggest that MoRFs are similar in composition to general intrinsically disordered proteins. However, several biases are inconsistent with this simple explanation. MoRFs are depleted or show similar composition to PDBS25 in charged residues other than arginine, which are generally disorder promoting. On the other hand, MoRFs are enriched in cysteine and phenylalanine.
Hub proteins of C. elegans PPI network
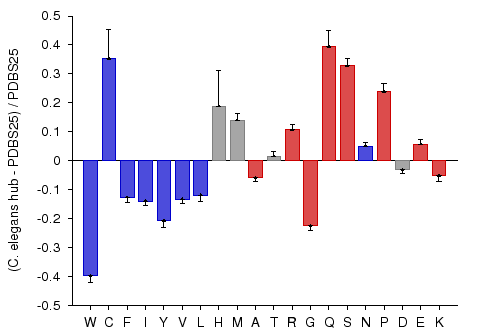
A potential association between protein connectivity and protein intrinsic disorder was studied for proteins with various numbers of interacting partners from four eukaryotic organisms (C. elegans, S. cerevisiae, D. melanogaster, and H. sapiens) (Haynes et al., 2006). A more detailed analysis revealed that hub proteins, defined as proteins interacting with at least 10 partners, are significantly more disordered than end proteins, defined as those that interact with just one partner. To test the compositional bias of hubs and ends, the fractional difference between hubs and ends compositions and PDBS25 compositions was calculated. This analysis revealed that that hubs are enriched in many of the disorder-promoting amino acids, whereas compositions of ends were shown to be relatively close to that of ordered proteins. This study demonstrated that intrinsic disorder is a distinctive and common characteristic of eukaryotic hub proteins, and that disorder may serve as a determinant of protein interactivity. This particular example shows the composition profile of hub proteins from C. elegans. The red-colored bars on the right hand side of the graph represent disorder-promoting residues.
References
- Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac
P, Uversky VN, Vidal M, Iakoucheva LM. (2006) "Intrinsic disorder is a
common feature of hub proteins from four eukaryotic interactomes."
PLoS Comput Biol. 2
(8):e100.

- Janin J, Miller S, and Chothia C. (1988) "Surface, subunit
interfaces and interior of oligomeric proteins." J Mol Biol,
204:155-64.

- Janin J and Chothia C. (1990) "The structure of protein-protein
recognition sites." J Biol Chem, 265:16027-30.

- Jones S and Thornton JM. (1996) "Principles of protein-protein
interactions." Proc Natl Acad Sci USA, 93:13-20.

- Jones S and Thornton JM. (1997) "Analysis of protein-protein
interaction sites using surface patches." J Mol Biol. 272
(1):121-32.

- Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK,
and Uversky VN. (2006) "Analysis of molecular recognition features
(MoRFs)." J Mol Biol. 362(5):1043-59.

- Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, and Dunker
AK. (2005) "Coupled folding and binding with alpha-helix-forming
molecular recognition elements." Biochemistry. 44
(37):12454-70.

- Valdar WS and Thornton JM. (2001) "Protein-protein interfaces:
analysis of amino acid conservation in homodimers." Proteins,
42:108-24.
